5
Import Data
Tidy Data Science with the Tidyverse and Tidymodels
W. Jake Thompson
https://tidyds-2021.wjakethompson.com · https://bit.ly/tidyds-2021
Tidy Data Science with the Tidyverse and Tidymodels is licensed under a Creative Commons Attribution 4.0 International License.
Incredibly boring, or...
absolutely infuriating.
Your turn 1
02:00
- Open the R Notebook materials/exercises/05-import.Rmd
- Run the setup chunk
Your turn 1
02:00
- Open the R Notebook materials/exercises/05-import.Rmd
- Run the setup chunk

Be kind to your collaborators...
Be kind to your collaborators...
including future you.
Be kind to your collaborators...
including future you.
Workflow
- Editor
- Home directory
- R code you ran before break
Be kind to your collaborators...
including future you.
Workflow
- Editor
- Home directory
- R code you ran before break
Product
- Raw data
- R code someone else need to run to replicate results
Be kind to your collaborators...
including future you.
Workflow
- Editor
- Home directory
- R code you ran before break
Product
- Raw data
- R code someone else need to run to replicate results
- Workflows should not be hardwired into the products
Projects
Each analysis as a project
- Folder on your computer with all relevant files
R scripts written with assumption of:
- Clean session
- Working directory = project directory
Creates everything it needs, touches nothing it didn't create.
Can move directory on computer, can move to different computer, can be used by other person (including future you!)
It’s like agreeing that we will all drive on the left or the right. A hallmark of civilization is following conventions that constrain your behavior a little, in the name of public safety.
Artwork by @allison_horst
here()
Find the project directory and build file paths.
library(here)here()#> [1] "/Users/jakethompson/Documents/GIT/courses/tidyds-2021"here("materials", "data", "nimbus.csv")#> [1] "/Users/jakethompson/Documents/GIT/courses/tidyds-2021/materials/data/nimbus.csv"here()
Where does here() start?
Is a file named
.herepresent?Is there a
.Rprojfile (e.g.,tidyds-2021.Rproj)?Is there a
.gitor.svndirectory?
dr_here()#> here() starts at /Users/jakethompson/Documents/GIT/courses/tidyds-2021.#> - This directory contains a file matching "[.]Rproj$" with contents matching "^Version: " in the first line#> - Initial working directory: /Users/jakethompson/Documents/GIT/courses/tidyds-2021/site/static/slides#> - Current working directory: /Users/jakethompson/Documents/GIT/courses/tidyds-2021/site/static/slides(Applied) Data Science
readr functions
| function | extracts |
|---|---|
| read_csv() | comma separated files |
| read_csv2() | semi-colon separated files |
| read_delim() | general delimited files |
| read_fwf() | fixed width files |
| read_log() | Apache log files |
| read_table() | space separated files |
| read_tsv() | tab separated files |
readr functions
| function | extracts |
|---|---|
| read_csv() | comma separated files |
| read_csv2() | semi-colon separated files |
| read_delim() | general delimited files |
| read_fwf() | fixed width files |
| read_log() | Apache log files |
| read_table() | space separated files |
| read_tsv() | tab separated files |
Example data: nimbus
#> date,longitude,latitude,ozone#> 1985-10-01T00:00:00Z,-179.375,-73.5,302#> 1985-10-01T00:00:00Z,-178.125,-73.5,302#> 1985-10-01T00:00:00Z,-176.875,-73.5,302#> 1985-10-01T00:00:00Z,-175.625,-73.5,302#> 1985-10-01T00:00:00Z,-174.375,-73.5,304#> 1985-10-01T00:00:00Z,-173.125,-73.5,304#> 1985-10-01T00:00:00Z,-171.875,-73.5,304#> 1985-10-01T00:00:00Z,-170.625,-73.5,304#> 1985-10-01T00:00:00Z,-164.375,-73.5,287read_csv()
readr functions share a common syntax.
dat <- read_csv("path/to/file.csv", ...)
read_csv()
readr functions share a common syntax.
dat <- read_csv("path/to/file.csv", ...)
object to save data to
read_csv()
readr functions share a common syntax.
dat <- read_csv("path/to/file.csv", ...)
read_csv()
readr functions share a common syntax.
dat <- read_csv(here("path", "to", "file.csv"), ...)
build file path with here()
Your turn 2
Find
nimbus.csvin your project directoryRead it into an object
View the results
02:00
nimbus <- read_csv(here("materials", "data", "nimbus.csv"))#> #> ── Column specification ────────────────────────────────────────────────────────#> cols(#> date = col_datetime(format = ""),#> longitude = col_double(),#> latitude = col_double(),#> ozone = col_character()#> )nimbus#> # A tibble: 18,963 x 4#> date longitude latitude ozone#> <dttm> <dbl> <dbl> <chr>#> 1 1985-10-01 00:00:00 -179. -73.5 302 #> 2 1985-10-01 00:00:00 -178. -73.5 302 #> 3 1985-10-01 00:00:00 -177. -73.5 302 #> 4 1985-10-01 00:00:00 -176. -73.5 302 #> 5 1985-10-01 00:00:00 -174. -73.5 304 #> 6 1985-10-01 00:00:00 -173. -73.5 304 #> 7 1985-10-01 00:00:00 -172. -73.5 304 #> 8 1985-10-01 00:00:00 -171. -73.5 304 #> 9 1985-10-01 00:00:00 -164. -73.5 287 #> 10 1985-10-01 00:00:00 -163. -73.5 287 #> # … with 18,953 more rowstibbles
read.csv() vs. read_csv()
#> #> 18939 1985-10-01T00:00:00Z 139.375 -0.5 270#> 18940 1985-10-01T00:00:00Z 140.625 -0.5 275#> 18941 1985-10-01T00:00:00Z 141.875 -0.5 270#> 18942 1985-10-01T00:00:00Z 143.125 -0.5 266#> 18943 1985-10-01T00:00:00Z 144.375 -0.5 267#> 18944 1985-10-01T00:00:00Z 145.625 -0.5 263#> 18945 1985-10-01T00:00:00Z 146.875 -0.5 261#> 18946 1985-10-01T00:00:00Z 148.125 -0.5 262#> 18947 1985-10-01T00:00:00Z 154.375 -0.5 271#> 18948 1985-10-01T00:00:00Z 155.625 -0.5 272#> 18949 1985-10-01T00:00:00Z 156.875 -0.5 268#> 18950 1985-10-01T00:00:00Z 158.125 -0.5 276#> 18951 1985-10-01T00:00:00Z 159.375 -0.5 273#> 18952 1985-10-01T00:00:00Z 160.625 -0.5 272#> 18953 1985-10-01T00:00:00Z 161.875 -0.5 271#> 18954 1985-10-01T00:00:00Z 163.125 -0.5 272#> 18955 1985-10-01T00:00:00Z 164.375 -0.5 275#> 18956 1985-10-01T00:00:00Z 165.625 -0.5 271#> 18957 1985-10-01T00:00:00Z 166.875 -0.5 271#> 18958 1985-10-01T00:00:00Z 168.125 -0.5 273#> 18959 1985-10-01T00:00:00Z 169.375 -0.5 273#> 18960 1985-10-01T00:00:00Z 170.625 -0.5 271#> 18961 1985-10-01T00:00:00Z 171.875 -0.5 270#> 18962 1985-10-01T00:00:00Z 173.125 -0.5 268#> 18963 1985-10-01T00:00:00Z 174.375 -0.5 265read.csv() vs. read_csv()
#> #> 18939 1985-10-01T00:00:00Z 139.375 -0.5 270#> 18940 1985-10-01T00:00:00Z 140.625 -0.5 275#> 18941 1985-10-01T00:00:00Z 141.875 -0.5 270#> 18942 1985-10-01T00:00:00Z 143.125 -0.5 266#> 18943 1985-10-01T00:00:00Z 144.375 -0.5 267#> 18944 1985-10-01T00:00:00Z 145.625 -0.5 263#> 18945 1985-10-01T00:00:00Z 146.875 -0.5 261#> 18946 1985-10-01T00:00:00Z 148.125 -0.5 262#> 18947 1985-10-01T00:00:00Z 154.375 -0.5 271#> 18948 1985-10-01T00:00:00Z 155.625 -0.5 272#> 18949 1985-10-01T00:00:00Z 156.875 -0.5 268#> 18950 1985-10-01T00:00:00Z 158.125 -0.5 276#> 18951 1985-10-01T00:00:00Z 159.375 -0.5 273#> 18952 1985-10-01T00:00:00Z 160.625 -0.5 272#> 18953 1985-10-01T00:00:00Z 161.875 -0.5 271#> 18954 1985-10-01T00:00:00Z 163.125 -0.5 272#> 18955 1985-10-01T00:00:00Z 164.375 -0.5 275#> 18956 1985-10-01T00:00:00Z 165.625 -0.5 271#> 18957 1985-10-01T00:00:00Z 166.875 -0.5 271#> 18958 1985-10-01T00:00:00Z 168.125 -0.5 273#> 18959 1985-10-01T00:00:00Z 169.375 -0.5 273#> 18960 1985-10-01T00:00:00Z 170.625 -0.5 271#> 18961 1985-10-01T00:00:00Z 171.875 -0.5 270#> 18962 1985-10-01T00:00:00Z 173.125 -0.5 268#> 18963 1985-10-01T00:00:00Z 174.375 -0.5 265#> # A tibble: 18,963 x 4#> date longitude latitude ozone#> <dttm> <dbl> <dbl> <chr>#> 1 1985-10-01 00:00:00 -179. -73.5 302 #> 2 1985-10-01 00:00:00 -178. -73.5 302 #> 3 1985-10-01 00:00:00 -177. -73.5 302 #> 4 1985-10-01 00:00:00 -176. -73.5 302 #> 5 1985-10-01 00:00:00 -174. -73.5 304 #> 6 1985-10-01 00:00:00 -173. -73.5 304 #> 7 1985-10-01 00:00:00 -172. -73.5 304 #> 8 1985-10-01 00:00:00 -171. -73.5 304 #> 9 1985-10-01 00:00:00 -164. -73.5 287 #> 10 1985-10-01 00:00:00 -163. -73.5 287 #> # … with 18,953 more rowsparsing
Consider
Look at the
nimbusdata.What class (data type) is
ozone?
nimbus %>% pull(ozone) %>% class()01:00
nimbus %>% pull(ozone) %>% class()#> [1] "character"nimbus %>% pull(ozone) %>% unique()#> [1] "302" "304" "287" "274" "264" "242" "211" "195" "197" "196" "198" "193"#> [13] "187" "190" "199" "194" "213" "218" "221" "229" "209" "186" "188" "191"#> [25] "189" "184" "180" "." "215" "312" "319" "320" "311" "300" "290" "267"#> [37] "226" "210" "200" "203" "201" "192" "204" "206" "208" "205" "223" "232"#> [49] "238" "243" "220" "202" "185" "219" "222" "216" "324" "336" "333" "323"#> [61] "308" "295" "244" "212" "237" "248" "239" "241" "250" "249" "252" "234"#> [73] "318" "313" "326" "335" "337" "316" "266" "207" "227" "251" "253" "257"#> [85] "261" "214" "228" "273" "285" "288" "291" "270" "254" "317" "325" "332"#> [97] "340" "344" "338" "297" "247" "217" "225" "231" "235" "236" "262" "260"#> [109] "265" "272" "278" "280" "279" "255" "245" "224" "181" "240" "269" "296"#> [121] "307" "315" "321" "306" "299" "298" "283" "327" "322" "328" "331" "310"#> [133] "275" "233" "258" "276" "281" "289" "330" "346" "305" "334" "359" "347"#> [145] "314" "301" "256" "263" "277" "284"#> [ reached getOption("max.print") -- omitted 82 entries ]NA values
nimbus %>% filter(ozone == ".")#> # A tibble: 155 x 4#> date longitude latitude ozone#> <dttm> <dbl> <dbl> <chr>#> 1 1985-10-01 00:00:00 70.6 -73.5 . #> 2 1985-10-01 00:00:00 71.9 -73.5 . #> 3 1985-10-01 00:00:00 73.1 -73.5 . #> 4 1985-10-01 00:00:00 74.4 -73.5 . #> 5 1985-10-01 00:00:00 75.6 -73.5 . #> 6 1985-10-01 00:00:00 76.9 -73.5 . #> 7 1985-10-01 00:00:00 78.1 -73.5 . #> 8 1985-10-01 00:00:00 79.4 -73.5 . #> 9 1985-10-01 00:00:00 65.6 -72.5 . #> 10 1985-10-01 00:00:00 66.9 -72.5 . #> # … with 145 more rowsDefine missing values
dat <- read_csv(here("path", "to", "file.csv"), na = ".")
Your turn 3
Read in
nimbus.csvagain.Set values of
"."toNA.
02:00
read_csv(here("materials", "data", "nimbus.csv"))#> #> ── Column specification ────────────────────────────────────────────────────────#> cols(#> date = col_datetime(format = ""),#> longitude = col_double(),#> latitude = col_double(),#> ozone = col_character()#> )#> # A tibble: 18,963 x 4#> date longitude latitude ozone#> <dttm> <dbl> <dbl> <chr>#> 1 1985-10-01 00:00:00 -179. -73.5 302 #> 2 1985-10-01 00:00:00 -178. -73.5 302 #> 3 1985-10-01 00:00:00 -177. -73.5 302 #> 4 1985-10-01 00:00:00 -176. -73.5 302 #> 5 1985-10-01 00:00:00 -174. -73.5 304 #> 6 1985-10-01 00:00:00 -173. -73.5 304 #> 7 1985-10-01 00:00:00 -172. -73.5 304 #> 8 1985-10-01 00:00:00 -171. -73.5 304 #> 9 1985-10-01 00:00:00 -164. -73.5 287 #> 10 1985-10-01 00:00:00 -163. -73.5 287 #> # … with 18,953 more rowsread_csv(here("materials", "data", "nimbus.csv"), na = ".")#> #> ── Column specification ────────────────────────────────────────────────────────#> cols(#> date = col_datetime(format = ""),#> longitude = col_double(),#> latitude = col_double(),#> ozone = col_double()#> )#> # A tibble: 18,963 x 4#> date longitude latitude ozone#> <dttm> <dbl> <dbl> <dbl>#> 1 1985-10-01 00:00:00 -179. -73.5 302#> 2 1985-10-01 00:00:00 -178. -73.5 302#> 3 1985-10-01 00:00:00 -177. -73.5 302#> 4 1985-10-01 00:00:00 -176. -73.5 302#> 5 1985-10-01 00:00:00 -174. -73.5 304#> 6 1985-10-01 00:00:00 -173. -73.5 304#> 7 1985-10-01 00:00:00 -172. -73.5 304#> 8 1985-10-01 00:00:00 -171. -73.5 304#> 9 1985-10-01 00:00:00 -164. -73.5 287#> 10 1985-10-01 00:00:00 -163. -73.5 287#> # … with 18,953 more rowsSpecify column types
dat <- read_csv(here("path", "to", "file.csv"), na = ".",
col_types = cols(var_1 = col_number()))
Column types
| function | data type |
|---|---|
| col_character() | characters |
| col_date() | dates |
| col_datetime() | POSIXct (date-time) |
| col_double() | double (decimal number) |
| col_factor() | factors |
| col_guess() | let readr guess (default) |
| col_integer() | integers |
| col_logical() | logicals |
| col_number() | numbers mixed with non-number characters |
| col_numeric() | double or integer |
| col_skip() | do not read |
| col_time() | time |
Your turn 4
Read in
nimbus.csvagain.Set values of
"."toNA.Specify
ozoneas integer values.
02:00
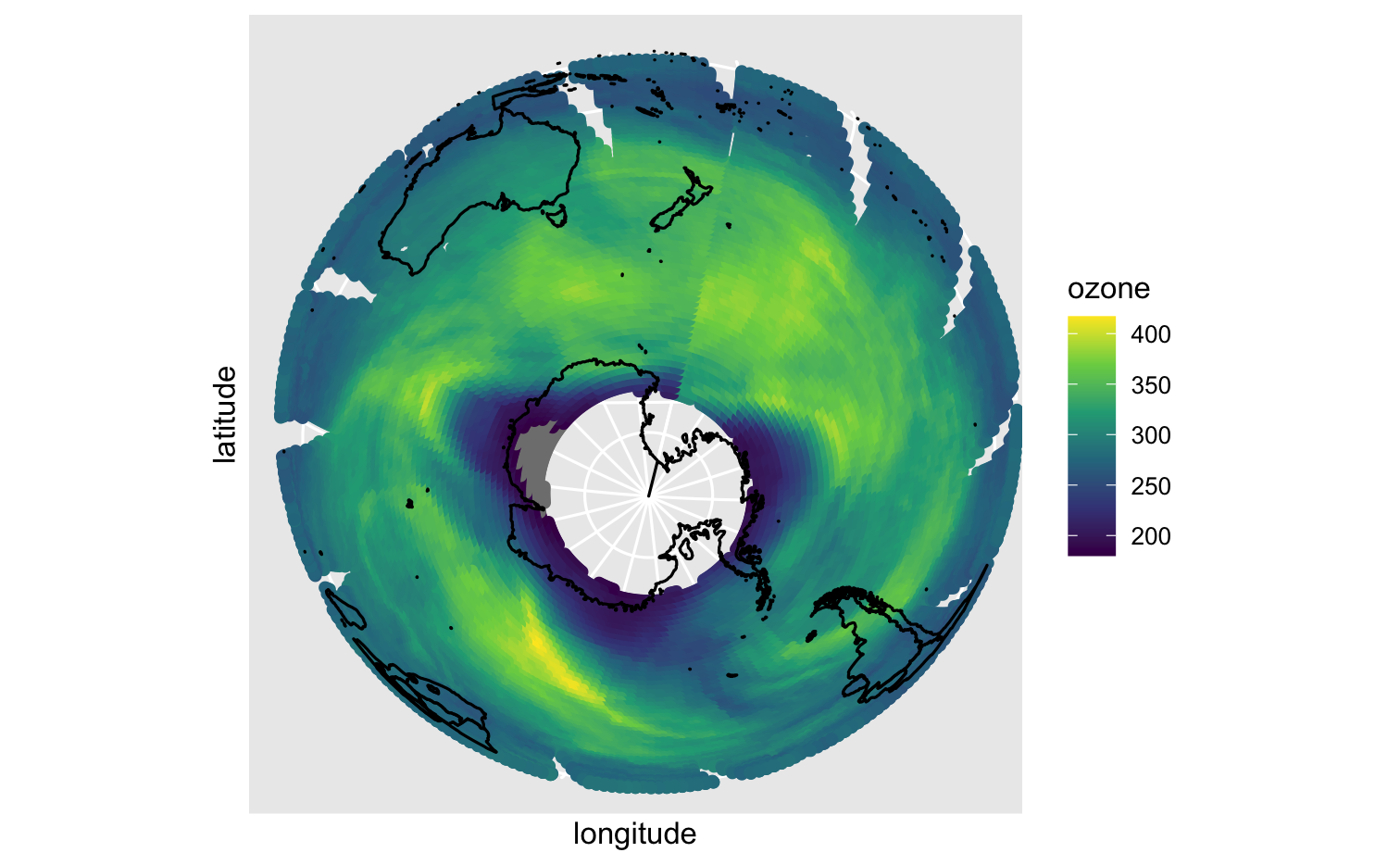
read_csv(here("materials", "data", "nimbus.csv"), na = ".", col_types = cols(ozone = col_integer()))#> # A tibble: 18,963 x 4#> date longitude latitude ozone#> <dttm> <dbl> <dbl> <int>#> 1 1985-10-01 00:00:00 -179. -73.5 302#> 2 1985-10-01 00:00:00 -178. -73.5 302#> 3 1985-10-01 00:00:00 -177. -73.5 302#> 4 1985-10-01 00:00:00 -176. -73.5 302#> 5 1985-10-01 00:00:00 -174. -73.5 304#> 6 1985-10-01 00:00:00 -173. -73.5 304#> 7 1985-10-01 00:00:00 -172. -73.5 304#> 8 1985-10-01 00:00:00 -171. -73.5 304#> 9 1985-10-01 00:00:00 -164. -73.5 287#> 10 1985-10-01 00:00:00 -163. -73.5 287#> # … with 18,953 more rowslibrary(rnaturalearth)library(sf)world <- ne_countries(scale = "medium", returnclass = "sf")ortho <- "+proj=ortho +lat_0=-78 +lon_0=166 +x_0=0 +y_0=0 +a=6371000 +b=6371000 +units=m +no_defs"ggplot(data = nimbus) + geom_point(mapping = aes(x = longitude, y = latitude, color = ozone)) + geom_sf(data = world, fill = NA, color = "black") + scale_color_viridis_c(option = "viridis") + coord_sf(crs = ortho)
other data files
Excel files (.xls and .xlsx)

Data from other statistical software (SPSS, Stata, and SAS)

Google Sheets and other files from Google Drive

Efficient data sharing between R and Python

Web pages (web scraping)

jsonlite -> json xml2 -> xml httr -> web APIs DBI -> databases
Import Data

Tidy Data Science with the Tidyverse and Tidymodels
W. Jake Thompson
https://tidyds-2021.wjakethompson.com · https://bit.ly/tidyds-2021
Tidy Data Science with the Tidyverse and Tidymodels is licensed under a Creative Commons Attribution 4.0 International License.